News
News & blog posts

Processing rMATS output
May 25, 2023
rMATS outputs several files, including txt files for all considered splicing events (i.e. SE (skipped exon), MXE (mutually exclusive exons),...

Alternative splicing and Snakemake
May 18, 2023
In this and next few posts, I will log my attempt to replace shell scripting (which I have been using...

The lab has grown!
December 20, 2022
In the autumn of 2022 we have welcomed two new lab members. Bogna joined as a lab manager and Ola...
TT-seq analysis -Spliced reads
December 20, 2022
In this post, I will continue with reproducing the analysis from Shao et al and compare spliced reads across...
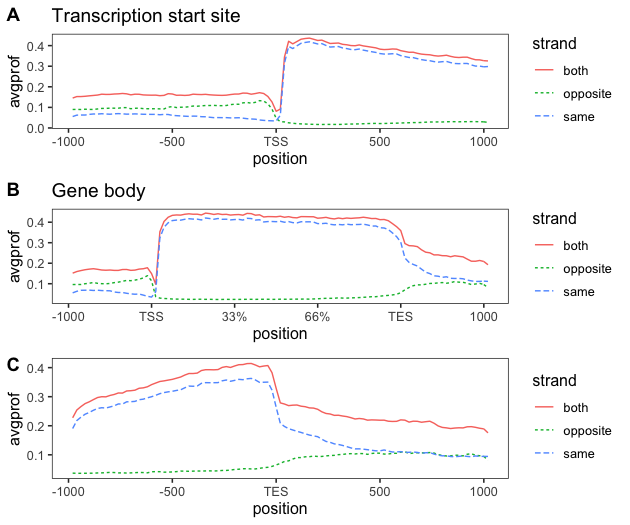
TT-seq analysis - Metagene profiles 2
September 22, 2022
In this post, I will re-plot the graphs created using ngsplot, to combine the sense and anti-sense profiles on a...
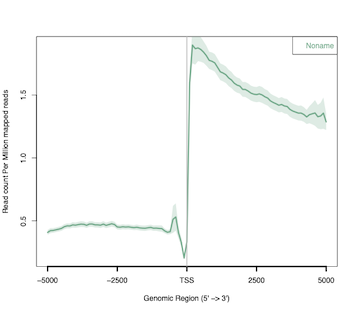
TT-seq analysis - Metagene profiles
September 15, 2022
In this post, I will use ngs.plot to visualize the results of TT-seq experiment around functional genomic regions, i.e. transcription...
TT-seq analysis - TU annotation
August 18, 2022
In this post, we will use R shiny application developed by Shao et al. (1), to annotate transcriptional units in...

New lab space
August 17, 2022
We moved to the Centre of Advanced Technologies. It is not far - geographically speaking, but it is a big...

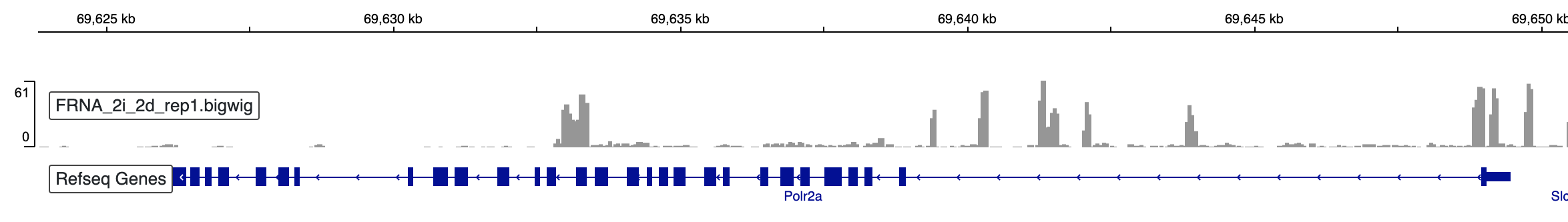
TT-seq analysis - BigWig files
July 07, 2022
In this post, we will use previously calculated scale factors to create scale and strand-specific BigWig files. These files can...
TT-seq analysis - Scale factors
June 23, 2022
In this post, I will normalize the sequencing samples, using the read counts from ERCC spike-ins. To this end I...

TT-seq analysis - Step 1
June 16, 2022
The data I have been working with are the result of TT-seq experiment performed by Shao et al., who set...

Reads alignment
May 26, 2022
Today, I will start the analysis of the data and align the pair-end reads to mm10 reference sequence using STAR.
...
Downloading fastq files.
May 19, 2022
Let’s pick it up where we left it last time. SRA explorer provides as with basic bash script that we...

Accessing and downloading raw sequencing data.
April 21, 2022
GEO does not store raw data. As confusing as it seems, GEO does not store raw sequence data. Various formats...

Downloading data from GEO to HPC.
April 14, 2022
Most, if not all (published) NGS data is deposited to GEO repository and available to download and process. Today the...

Bash profile
April 07, 2022
We will be using shell to do most of our work. Shell accepts and runs commands. When you login, few...

Welcome to Maslon Lab
March 24, 2022
Welcome to our website. This space will be used to advertise new positions, talk about our research and share my...
